Is Genetic Makeup Or Allele Combination
Introduction
Climate change has begun to negatively impact yield of cereal crops (Lesk et al., 2016) and is predicted to cause fifty-fifty further yield losses in many of the low latitude grain producing regions of the earth (Dai, 2011; Rosenzweig et al., 2014). At the same fourth dimension global demand for cereals keeps growing (Tilman et al., 2011) caused by both population increment (Gerland et al., 2014) and altered consumption patterns (Kastner et al., 2012). The Nordic region is unique from an agricultural perspective with its relatively mild climate for its northern latitude and a long photoperiod during the growth season (Nurminiemi et al., 1996). Barley (Hordeum vulgare L.) is, alongside wheat, one of the dominating cereal crops in the Nordic region1, used primarily for feed and malt but with a growing need for human consumption (Baik and Ullrich, 2008; Baik, 2016). Recent breeding efforts have paved the way for a more than reliable barley harvest in the northern marginal area (Lillemo et al., 2010; Hilmarsson et al., 2017), where early flowering and the ability to reach maturity at low temperatures are fundamental components to secure a loftier and stable yield at high latitudes (Nurminiemi et al., 1996; Hilmarsson et al., 2017). Events of strong winds and heavy precipitation are likely to increase in frequency due to global warming (Coumou and Rahmstorf, 2012). Hence, resistance to lodging and straw breaking are important traits. The oestrus wave in Scandinavia in the summertime of 2018, which led to considerable yield losses, further stresses the importance to address a more volatile and unpredictable future climate. Hither, early developing cultivars, which need less time in the field and are thus exposed to potentially damaging weather for a shorter time, could play a office in mitigating the risks. Amend agreement of the genetics underlying these traits will enable breeders to produce locally adapted loftier yielding cultivars for the Nordic and sub-arctic region, farther expanding the current cultivation expanse northwards.
Timing of flowering through seasonal cues, such as day length and temperature, is a key chemical element for reproductive success (Andrés and Coupland, 2012). Earliness is a complex trait where genetic variation can greatly alter the plants response to photoperiod and temperature (Cockram et al., 2007; Blümel et al., 2015). In its region of origin, barley germinates in the fall and stays in the vegetative phase during the cool and humid winter season; increased day length in the bound triggers the onset of flowering and the plants mature at the start of the dry out summer period ensuring a period of dormancy for the seeds during the hot and dry summertime (Lister et al., 2009). Consistent with its importance for the institute'south survival the response to changes in the photoperiod is controlled past several well conserved genes (Blümel et al., 2015). Among those is the Ppd-H1 gene, located on chromosome 2H, whose wild type function is to promote flowering nether long day atmospheric condition (Turner et al., 2005; Jones et al., 2008; Loscos et al., 2014). With the expansion of barley northward with the spread of agriculture, a recessive ppd-H1 allele with delayed flowering was favored (Jones et al., 2008). This recessive allele helped the spring barley utilize the summertime season in the northern latitudes past a less strong upwards-regulation of the HvFT1 gene than with the wild type Ppd-H1 allele (Hemming et al., 2008). Lister et al. (2009) found a latitudinal increase in the prevalence of this recessive allele in historical cultivars and landraces from Europe. Another of import gene for the regulation of flowering is HvCO1, a gene interim in parallel with Ppd-H1, whose overexpression leads to up-regulation of the HvFT1 gene, which in plough leads to flowering (Campoli et al., 2012; Loscos et al., 2014). HvCO2 on chromosome 6H is a paralog to HvCO1 (Campoli et al., 2012). HvFT1 has alleles with copy number variation (CNV) which have been associated with early flowering in spring barley, a phenotype offset discovered in the Finnish cultivar Tammi (Nitcher et al., 2013; Loscos et al., 2014). HvFT1 has two paralogs: Ppd-H2 (synonym HvFT3) which promotes spikelet initiation (Mulki et al., 2018) and HvCEN, which has been shown to touch flowering time and has a latitudinal specific distribution of alleles suggesting adaptive part in the northwards range expansion of barley (Comadran et al., 2012). The HvCEN (syn. eps2S or eam6; Comadran et al., 2012; Alqudah et al., 2016) locus inhibits flowering and is located in the centromeric region of chromosome 2H (Comadran et al., 2012). HvCEN has been shown to have a mutant allele that, contrary to the wild type allele, does non inhibit flowering in spring barley and interact with HvFT1 (Loscos et al., 2014). Another FT-like gene is HvFT4 on the brusk arm of chromosome 2H which is a temperature responsive gene with increased expression in loftier temperature (Ford et al., 2016). HvELF3 (syn. Mat-a or Eam8) on chromosome 1H is a homolog of Arabidopsis thaliana factor ELF3 (Zakhrabekova et al., 2012). The dominant HvELF3 allele delays flowering in long day conditions while several of the recessive alleles provide mean solar day length neutrality which leads to early flowering in both long-day and brusk-twenty-four hour period weather (Faure et al., 2012). I recessive allele (mat-a.eight) is the result of an induced mutation in the cultivar Bonus and was released 1960 with the cultivar Mari (Gustafsson et al., 1971; Lundqvist, 2009), the name describing its main characteristics (from Latin for matura = early on and rigida = stiff) (Gustafsson et al., 1971). The twenty-four hours length neutrality associated with the HvELF3 polymorphism has been proposed to enable cultivation of barley as far north as Iceland, equally well as enabling the spread of barley to loftier altitude regions near the equator (Faure et al., 2012; Zakhrabekova et al., 2012). Vrn-H1 (HvAP1) on chromosome 5H is involved in vernalization requirement and interacts with Vrn-H2 on chromosome 4H. A wild type recessive vrn-H1 and a functional Vrn-H2 e'er result in a winter growth habit (Karsai et al., 2005; Loscos et al., 2014). Several alleles of Vrn-H1 exists, resulting in a bound growth habit or a facultative growth habit in lines with a jump allele in Vrn-H1 or lines where Vrn-H2 is deleted (Loscos et al., 2014).
In add-on to the importance of the correct timing of flowering, plant height matters every bit demonstrated when dwarfing genes were introduced into wheat in the green revolution (Peng et al., 1999). Intensive cereal cultivation is today dependent on semi-dwarf cultivars (Kuczyńska et al., 2013) since short and strong stems help the plants withstand wind, foreclose lodging and can positively impact the harvest index (Hay, 1995). Studies have shown that height reduction is the result of either reduced hormone expression or hormone insensitivity (Dockter et al., 2014), and that two of the main factors regulating institute height are the plant hormones brassinosteroids (BRs) and gibberellic acids (GAs) (Marzec and Alqudah, 2018). BRs are as well known to affect traits such as tiller number and grain size in rice (Zhang et al., 2014). In barley, height is controlled by dwarfing and semi-dwarfing genes every bit well as other genes affecting institute peak (Wang et al., 2014). The dwarfing genes are not useful in breeding every bit they are linked to reduced vigor and yield (Wang et al., 2014). Instead, semi-dwarfing genes have been widely employed in modern barley breeding (Kuczyńska et al., 2013; Wang et al., 2014), these include semi-brachytic 1 (uzu1) (Chono et al., 2003), semi-dwarf i (sdw1/denso) (Jia et al., 2009), breviaristatum-e (ari-e) (Liu et al., 2014), and short culm ane (hcm1) (Wang et al., 2014). The uzu1 and sdw1/denso genes are both located close to the centromere on chromosomal arm 3HL with the sdw1/denso cistron located more distally from the centromere. The ari-e locus is located on chromosomal arm 5HL and the hcm1 gene is located on chromosomal arm 2HL (Wang et al., 2014). Modern European barley cultivars more often than not depend on the sdw1/denso locus as their source of semi-dwarfing (Kuczyńska et al., 2013). Plants carrying the semi-dwarf allele of the sdw1/denso locus can be identified morphologically past having a prostrate growth addiction in their juvenile phase, whereas plants carrying the ascendant allele have an cock juvenile growth habit (Kuczyńska et al., 2013).
Loftier-throughput genotyping has developed every bit a viable culling to traditional genotyping with molecular markers, such every bit AFLPs. The high-throughput method utilizes single nucleotide polymorphisms (SNP) spread beyond the genome at an even distribution (Comadran et al., 2012). The evolution of high-throughput SNP-panels enables a genomic resolution not easily obtained by other marker types. This improved coverage has opened upward the possibility to perform genome wide association scans (GWAS) on a variety of agricultural traits (see e.g., Waugh et al., 2014).
Linkage disequilibrium (LD) is the non-random co-segregation of alleles at ii loci (Flintstone-Garcia et al., 2003). LD is by and large higher in self-pollinating crops than in out-convenance species and is higher in homogenous than in diverse populations (Flint-Garcia et al., 2003). In clan mapping, LD affects the number of markers needed as well as the resolution obtained in the associations (Rafalski, 2002). In self-pollinating crops with very high LD the resolution gets lower every bit associated markers may be located far away from the responsible locus (Malysheva-Otto et al., 2006). In contrast, when LD is low, the resolution is high as the distance between associated marker and the gene of interest will be short (Remington et al., 2001). A recent study of the same Nordic population as used here confirmed the boilerplate LD to be in the range 0–4 cM but with large variations in unlike chromosomal regions and varying among the population construction groups (Bengtsson et al., 2017).
Ameliorate understanding of the allelic diversity in the Nordic convenance material will enable the application of marker-assisted option for beneficial allele combinations and speed up the breeding process. Detailed agreement of loci decision-making earliness and straw stability will enable fine-tuning of cultivars better adapted to northern latitudes. We screened a panel of 169 barley lines from the Nordic breeding puddle at 8 locations for the traits heading day, maturity twenty-four hours and straw stability with the aim of performing a genome-wide association assay to identify loci responsible for the quantitative traits earliness and harbinger stability in multiple environments.
Materials and Methods
Plant Textile, Test Locations, and Phenotyping
A panel of 169 spring barley lines, representing the Nordic convenance gene puddle, were included in the written report, selected by each of six different Nordic barley breeding entities (Supplementary Table S1). The chief aim of the selection procedure was to maximize diversity for pathogen resistance, earliness and straw quality, among advanced cultivars and breeding lines. Out of the 169 lines – 124 ii-rowed and 45 six-rowed – 58 lines were of Danish origin (all ii-rowed), 30 Swedish (28 two-rowed and ii six-rowed), xxx Norwegian (iii two-rowed and 27 six-rowed), 29 Finnish (all two-rowed), 21 Icelandic (5 ii-rowed and xvi six-rowed), and one from the United Kingdom (2-rowed). The two panels used in this study, 1 consisting of all 169 lines and i with only the 124 2-rowed lines, are referred to equally PPP169 and PPP124, respectively.
Multi-environment field trials (MET) were performed in eight locations for 2–three years, resulting in a maximum of 19 distinct environments (Effigy 1A and Supplementary Tabular array S1), ranging from Laberweinting, Germany in the south (48°48′vi″N) to Korpa, Iceland in the north (64°08′56″N) and Jokioinen, Finland in the eastward (23°29′54″E) to Korpa in the west (21°45′03″W). Within the environments there is great variation in hours of sunlight during the growth flow (Figure 1B) and the rut sum available (Figure 1C). The field trials were ready up with upwards to three replications in an alpha lattice design. Plot size varied between locations from row sowings up to 10 m2 field plots. Lodging and harbinger breaking were non recorded in row sowings, since it could accept yielded a different outcome compared with field plots.

Effigy 1. An overview of the multi-environs field trials (MET). (A) Field tests were performed at eight different locations in 6 different countries, ranging from Laberweinting, Germany in the southward to Korpa, Iceland in the north, for up to 3 years, resulting in 19 singled-out environments. The number of years is shown within circles. (B) The different day lengths between test locations, with the respective sowing days of the trials. (C) The accumulated heat sum (°C) from sowing solar day until maturity of the latest maturing line in the eight trials where maturity was screened. The highest and lowest value are shown in the graph (at Korpa, IS 2013 not all lines reached maturity).
Plants were phenotyped for early jump growth in four environments (measured as height in all lines when the starting time lines entered growth stage 31, 32, and 34: Ht31, Ht32, and Ht34, respectively) (Zadoks et al., 1974). The measurement was conducted from soil level up to a cardboard plate laid on meridian of the plants in the plot, as an estimator of the height. Heading twenty-four hours (HD) was recorded in 17 environments as number of days from sowing until half of the spike was visible in 50% of the plants in each plot (stage 53; Zadoks et al., 1974). Maturity day (Doc) was recorded in 7 environments as number of days from sowing until the peduncle beneath the spike turned yellow (estimate growth stage 89; Zadoks et al., 1974). Grain filling period (GFP) was calculated as the period from HD to Md (7 environments). Heat sums for HD (HSHD), MD (HSMD), and GFP (HSGFP) were calculated by adding the maximum daily temperature with the minimum daily temperature and dividing them past 2, and so adding the sums for each mean solar day of the respective period giving the growing degree-days (GDD), with 0°C used as base line temperature, pregnant only sums in a higher place 0°C were added. Straw length (StL) was measured upon maturity from soil level to beneath the spike in cm (eleven environments). Lodging (Ld), recorded in half-dozen environments and straw breaking (SB), recorded in 5 environments were recorded on a scale from 1 to 9 where ane means no lodging/straw breaking, five ways l% lodging/straw breaking and 9 means 100% lodging/straw breaking.
DNA Extraction and Genotyping
DNA was extracted using a standard CTAB protocol (Cetyl Trimethyl Ammonium Bromide) with DNA extraction kits as described earlier by Orabi et al. (2014). All lines were genotyped with the barley 9K iSelect SNP chip which contains 7842 SNP markers (Comadran et al., 2012). Genotyping was performed by TraitGenetics (Gatersleben, Germany).
6208 SNP markers were polymorphic >v% level and their concrete position (in base pairs) on the barley reference genome (Mascher et al., 2017) were retrieved using the online tool BARLEYMAP2 (Cantalapiedra et al., 2015).
Descriptive Statistics
All traits were continuous except for the discrete character row blazon. For the continuous characters, a normal distribution was expected. To check for deviations from the expected normal distribution, information for all trials and traits were plotted in distribution plots using Excel Add-In XLSTAT five. nineteen.2. Pearson correlations were calculated to depict the relationships betwixt traits and betwixt field trials using Microsoft Excel 2016.
Descriptive statistics for all environments, separately and combined, were computed with the psych software packet v. 1.8.12 (Revelle, 2018) using the R software (R Development Cadre Team, 2017), this included number of observations (north), mean, standard deviation (sd), median, median absolute deviation (mad), minimum (min), maximum (max), range, skew, kurtosis, and standard error (se).
Analysis of Variance (ANOVA)
To evaluate the relative contributions of genotype, environment, and genotype by environment interactions in the data set, each trait was analyzed with mixed linear modeling using PROC MIXED in SAS v. 9.4 (SAS Institute Inc.). This initial analysis assumed genotypes, environments, and the genotype past environment interactions to be fixed furnishings and replications and blocks within replications of the individual trials as random effects.
All-time linear unbiased estimates (BLUEs) to be used in the GWAS analyses were calculated using the lmer role in the "lme4" R bundle (Bates et al., 2015), assuming all furnishings, except the genotypic effects to be random. Phenotypic data beyond years were estimated every bit:
where y ijk is the kth ascertainment of the ith genotype in the jth environment, μ is the common intercept, Gi is the effect of the ith genotype, enj is the consequence of the jth surround, r(j)m is the effect of the kth replication in surround j, and eijk is the corresponding fault.
Population Structure
Population structure was evaluated with the software Structure v.two.iii.iv (Falush et al., 2007) and by master component analysis (PCA) using GenAlEx v.six.5.0.1 (Peakall and Smouse, 2006; Peakall and Smouse, 2012; as in Bengtsson et al., 2017).
For the STRUCTURE assay, the SNP genotype data was run 10 times with a burn-in menstruation of 9999 followed by 9999 iterations from Chiliad = i to K = 12. To identify the optimal number of genetic clusters (subpopulations), ΔK values were calculated as proposed by Evanno et al. (2005) using STRUCTURE HARVESTER five. 0.6.94 (accessed 25 Nov. 2015)three. The Structure analysis was previously run for the PPP169 panel in Bengtsson et al. (2017), but here an identical analysis for the PPP124 panel was run. An assay of molecular variance (AMOVA) was run in GenAlEx to check for variance amid the STRUCTURE groups.
Genome-Wide Association Assay
Genome-broad association analysis was performed using TASSEL five. 5.2.31 (Bradbury et al., 2007) and using the R package, Genome Association and Prediction Integrated Tool (GAPIT v. iii.0) (Lipka et al., 2012). The hapmap file of the SNP markers was filtered to exclude unsuccessful marking assays, monomorphic markers, and rare alleles (with less than 5% occurrence in the population). Unmapped SNP markers were assigned to an artificial chromosome to capture any associations with these markers.
After filtering 5710 SNP markers remained in the analysis of the PPP169 panel, or 73% of the total number of 7864 SNPs. When the PPP124 panel was filtered to remove markers below 5% polymorphism, 5037 SNP markers remained. Kinship matrices were constructed based on the filtered fix of markers using the scaled identity-by-state (IBS) method (Zhang et al., 2010) for both panels. The corresponding kinship matrices were and then used in subsequent mixed linear model (MLM) analyses (Yu et al., 2006; Zhang et al., 2010). To select the correct model and account for population structure we performed four associations using TASSEL: (1) General linear model (GLM) assay without including population structure in the model; (two) Efficient mixed model association (EMMA) using the kinship matrix with Q-values from STRUCTURE K = 2; (3) EMMA using kinship matrix with eigenvalues from the PCA analysis; and (iv) EMMA using simply the kinship matrix. In addition, the models in TASSEL were compared with the following models using GAPIT 5. three.0: (5) MLM using the van Raden kinship; and (six) MLM using the van Raden kinship with eigenvalues from the PCA analysis. The meaning allelic effect estimate is given in relation to the pocket-size allele in the GAPIT output. The MLM was run as optimum level and the P3D (estimated in one case) variance component estimation. Quantile-quantile (Q-Q) plots were created by comparison expected and observed chi-square values and Manhattan plots showing positions of associated markers beyond the genome were constructed for each trait using the R package CMplotiv. Quantile-quantile plots (Q-Q plots) were plotted to appraise the goodness of fit of the model for each trait. Large deviations from the expected distribution mean that the model does not fit the data. The models were evaluated trait-wise past comparing the Q-Q plots and checking the narrow sense heritability values, where a higher heritability indicated that the model had a higher predictive value.
GWAS was performed trait-wise on the calculated Blues from all trials with the PPP169 panel, likewise equally on the PPP124 panel which includes only the two-rowed lines. A threshold value was calculated to estimate a significance level for the association analysis. The Bonferroni method based on the total number of markers for each console and a significance level of 0.05 gave a -log10(p) = 5.00 for the PPP124 panel and -logten(p) = v.06 for the PPP169 console. This is a very stringent method (Gupta et al., 2014) considering that many of the markers are strongly linked. Thus, a suggestive threshold, before published past Duggal et al. (2008), allowing for ane false positive per genome scan was estimated past dividing 1 past the number of markers for each panel. This resulted in a suggestive threshold of -log10(p) = 3.70 for PPP124 and -logten(p) = 3.76 for PPP169, which is considered every bit the significance threshold for the marker-trait associations identified in this study. Both thresholds are indicated in the Manhattan plots.
The length of the quantitative trait loci (QTL) were decided by calculating the LD between the most significant markers at each intra-chromosomal locus using the TASSEL software. The r 2-values earlier reported for each chromosome and each panel (Bengtsson et al., 2017, Supplementary Tables S3) were used as threshold values for determining whether a QTL should exist regarded every bit distinct or not.
Allele Frequencies and Combinations
Allele combinations were constructed for the traits Ht34, HSHD, HSMD, StL, and SB in the PPP169 console. The most significantly mark-trait associated SNP marker was used for each QTL and for practical reasons, the total combination of SNPs used to construct the allele combinations was express to 3. The result of each allele combination with at to the lowest degree five observations (lines) was calculated based on BLUE values and the significance of the effects was tested using the lm () part in R. Allele frequencies were calculated for the SNP markers used in construction of the allele combinations, besides as previously described SNP markers for earliness traits (Comadran et al., 2012; Maurer et al., 2015).
Results
Descriptive Statistics and Distributions
Summary statistics were calculated for all nineteen environments, individually and combined (Supplementary Tables S2, S3), and frequency distributions plotted for all phenotypic traits (Supplementary Figure S1). For early vigor (Ht31, Ht32, Ht34), the distributions were generally right-skewed indicating mostly late genotypes in the two panels, with a few very early on developing lines. StL, SB, and Ld had normal distributions in both panels. For earliness traits (HD, HSHD, Physician, HSMD, GFP, and HSGFP) the distributions were left-skewed, indicating a few early on lines with the majority being later developing in both panels. The earliest genotypes were all six rowed, hence were not represented in the PPP124 panel.
Assay of Variance and Correlations
For all traits in both panels the genotype, environment, and genotype past environs interaction were significant (p < 0.05) except for environs event in Ht32 in the PPP124 console (Supplementary Table S4). The results showed that the event of genotype by far outweighed the genotype by surroundings interaction for all traits. Pearson correlations performed on overall ways for all 12 traits (Tabular array 1) showed that the earliness traits (HD, HSHD, Medico, HSMD, GFP, and HSGFP) were all positively correlated (p < 0.01), but with negative correlations to the straw properties and the early vigor (StL, SB, Ld, Ht31, Ht32, and Ht34), these were in plough positively intercorrelated.

Table 1. Pearson pairwise correlations of overall means for days from sowing to heading (HD), accumulated estrus sum from sowing to heading (HSHD), days from sowing to maturity (Dr.), accumulated heat sum from sowing to maturity (HSMD), grain filling period (registered as the number of days between heading and maturity) (GFP), and the accumulated heat sum in the grain filling catamenia (HSGFP), early vigor (measured as meridian at growth stage 31, 32, and 34 (Zadoks et al., 1974) (Ht31, Ht32, and Ht34), straw length (StL), straw breaking (SB), and lodging (Ld) for both panel PPP124 and PPP169.
Pearson correlations performed trait-wise between trials (Supplementary Table S5) showed significant correlation between trials for earliness traits and early vigor. Subsequently, these traits were analyzed as ways of all trials in the GWAS analyses. More variation was constitute for straw properties, where single trials were not correlated with the rest. In these cases, trials which had meaning correlation were analyzed every bit means, whereas trials without significant correlation were analyzed separately.
Population Structure
The Construction analysis for the PPP169 panel analyzed in Bengtsson et al. (2017) gave a maximum ΔK value at K = 2, where K1 comprised two-rowed lines from southern regions (mainly Finland, southern Sweden, and Denmark), K2 comprised the six-rowed lines and the admixed group was comprised of 2-rowed lines from Kingdom of norway, Iceland and northern Sweden. Here, STRUCTURE analysis of the PPP124 panel revealed the most variation for two subpopulations and AMOVA showed 38% of the genetic variation explained betwixt Chiliad groups. If the 2 STRUCTURE groups were farther subdivided co-ordinate to Tondelli et al. (2013) with an admixed group with less than 0.7 proportion of the genetic variation assigned to K1 or K2, respectively, these three groups explain 35% of the total variation. For applied purposes, the admixed genotypes were assigned to groups, either due north-western or south-eastern, based on knowledge of breeding entity.
Genome-Broad Clan Analysis
GWAS was run for 13 and 12 traits for the PPP169 and PPP124 panels, respectively. The best model, of the six models evaluated, was selected for each trait post-obit the criteria mentioned in material and methods (Supplementary Table S6). For all but two traits MLM with the van Raden kinship matrix, was used. For Ld in both panels, MLM with van Raden kinship matrix and eigenvalues from the PCA were used to business relationship for population structure. Results from analyses using MLM van Raden in GAPIT were reported, due to the resulting associations, where GAPIT in a few cases (e.g., for fasten type) yielded more peaks that passed the significance threshold and could be explained by known loci for the respective traits.
In total, for all 12 traits analyzed (excluding spike morphology), 108 meaning markers with known genetic position were found with 50 and 45 markers unique for the PPP169 and PPP124 panels, respectively (Supplementary Tables S7, S8). In total 23 and eleven QTL were constitute in the PPP169 and PPP124 panels, respectively. GWAS results are presented for nine out of twelve traits analyzed, that is for early on leap growth stages 31, 32, and 34 (Figure 2), StL, straw breaking and lodging (Figure 3), and heat sum heading, rut sum maturity, and estrus sum GFP (Effigy iv). Manhattan plots of spike morphology are shown for the PPP169 panel (Figure 5).
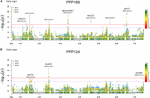
Effigy 2. GWAS results for the traits early spring growth stages 31, 32, and 34 (Zadoks et al., 1974), for panels (A) PPP169 and (B) PPP124, respectively. The Manhattan plots prove pregnant associations (Bonferroni threshold in red, and the suggestive threshold in purple) between trait and marking, the x-axis shows the physical altitude over all seven barley chromosomes. The bar under the x-centrality shows the SNP distribution on each chromosome, where 0 ->18 depicts SNP density (the number of SNPs per i Mbp bin). The not-pregnant and significant associations are displayed as open and solid filled circles, respectively. The significance of the associations at values between these two extremes are displayed as circles with varying amounts of fill. Relevant QTL names and putative loci are presented in the effigy. Note that the ∗ indicates a SNP mark located at 3H position 105 cM using the POPSEQ 2017 reference map (http://floresta.eead.csic.es/barleymap/).
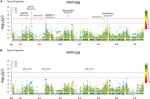
Figure 3. GWAS results for the traits straw length, straw breaking and lodging for panels (A) PPP169 and (B) PPP124, respectively. The Manhattan plots show significant associations (Bonferroni threshold in red, and the suggestive threshold in royal) between trait and marker, the x-axis shows the physical distance over all seven barley chromosomes. The bar nether the x-axis shows the SNP distribution on each chromosome, where 0 ->xviii depicts SNP density (the number of SNPs per 1 Mbp bin). The non-significant and significant associations are displayed as open up and solid filled circles, respectively. The significance of the associations at values between these two extremes are displayed every bit circles with varying amounts of fill. The most significant values, located at the top, are displayed as solid filled dots. The significance of the associations at values between these two extremes is indicated past all other dots with varying amounts of fill. Relevant QTL names and putative loci are presented in the figure.
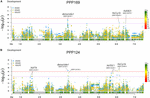
Figure iv. GWAS results for the traits estrus sum heading day, heat sum maturity, and heat sum grain filling menstruum for panels (A) PPP169 and (B) PPP124, respectively. The Manhattan plots evidence significant associations (Bonferroni threshold in red, and the suggestive threshold in royal) betwixt trait and marker, the 10-axis shows the concrete altitude over all vii barley chromosomes. The bar nether the x-axis shows the SNP distribution on each chromosome, where 0 ->18 depicts SNP density (the number of SNPs per 1 Mbp bin). The non-significant and meaning associations are displayed as open and solid filled circles, respectively. The significance of the associations at values betwixt these two extremes are displayed as circles with varying amounts of fill. The almost significant values, located at the tiptop, are displayed equally solid filled dots. The significance of the associations at values betwixt these two extremes is indicated by all other dots with varying amounts of make full. Relevant QTL names and putative loci are presented in the figure.

Effigy 5. GWAS results for spike morphology for console PPP169. The Manhattan plots prove significant associations (Bonferroni threshold in red, and the suggestive threshold in purple) between trait and marker, the 10-axis shows the concrete distance over all 7 barley chromosomes. The bar under the x-centrality shows the SNP distribution on each chromosome, where 0 ->xviii depicts SNP density (the number of SNPs per ane Mbp bin). The non-pregnant and significant associations are displayed every bit open and solid filled circles, respectively. The significance of the associations at values between these ii extremes are displayed equally circles with varying amounts of fill. The most significant values, located at the pinnacle, are displayed as solid filled dots. The significance of the associations at values between these 2 extremes is indicated by all other dots with varying amounts of fill. Relevant QTL names and putative loci are presented in the figure.
Allelic Diversity and Allele Combinations
Observed allelic diversity for selected SNP markers from the GWAS analyses, and for a set of SNP markers that accept previously been associated with flowering genes (Comadran et al., 2012; Maurer et al., 2015), showed unlike patterns of polymorphisms between the two geographic groups. Several loci were effectively fixed in the south-eastern lines, for example markers nearby or in the Ppd-H1, HvCO1, HvCO3, HvFT1, and denso/sdw1 loci (Supplementary Effigy S2). In contrast, alleles for markers nearby the Vrn-H1 locus were fixed in the north-western lines (Supplementary Effigy S2).
Allele combinations, with significance levels for the effects, were synthetic for the traits Ht34, HSHD, HSMD, StL, and SB in the PPP169 panel (Figure 6 and Supplementary Figure S3).
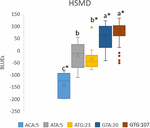
Figure half dozen. Boxplot of allele combinations showing Blues for rut sum to maturity (HSMD) (°C) in the PPP169 panel. Allele combinations were based on three significant markers that passed the suggestive threshold value in the GWAS analysis. The event of each allele combination for combinations with at least five observations (lines) was calculated based on Bluish values and the significance of the effects was tested using the lm () part in R. ∗ marks significant combinations (p ≤ 0.05). Allele combinations followed by the same lower example letter do non differ statistically.
Word
Latitudinal Adaptation of Earliness in Nordic Spring Barley
The barley lines analyzed here could be split up in two based on origin, that is into a due north-western group and a south-eastern grouping. The south-eastern grouping comprised 110 genotypes from breeders in Denmark, Finland, and southern Sweden. The due north-western group comprised 58 genotypes from breeders in northern Sweden, Norway, and Iceland. The unmarried genotype from the United Kingdom was not included in the geographical grouping. Despite the north-western group including fewer lines, more variety was observed at that place, possibly due to the fact that 6-rowed lines were almost exclusively found in this group. When looking simply at the PPP124 panel (all two-rows), the pattern was less clear.
A few of the well-known earliness loci had fixed alleles in the two-rowed lines, for instance markers in or nearby the Ppd-H1 and HvCEN locus, which could explain why these loci were non detected in the GWAS (Supplementary Figure S2). This is in accordance with previous findings that the wild type Ppd-H1 and HvCEN alleles are fixed in European ii-rowed spring barley (Tondelli et al., 2013). HvCEN has besides been reported as the locus well-nigh strongly associated with latitudinal issue among nineteen tested flowering associated loci in a European landrace drove (Russell et al., 2016).
Iii chromosomal regions take earlier been shown to be of importance in the regulation of flowering in barley; namely chromosomal arm 1HL (HvELF3 and Ppd-H2), the brusk and long arm of chromosome 2H (Ppd-H1 and HvCEN), and chromosomal arm 7HS (HvFT1 and HvCO1) (Loscos et al., 2014). Hither nosotros report associations on 1HL for early vigor in the PPP169 panel, simply not in the PPP124 console, with QDEV.169.1H-4 located only 4 Mbp (2.6 cM) from the HvELF3 locus (Figure 2A). QDEV.169.1H-iv showed diversity in the due south-eastern two-rowed lines merely had stock-still alleles in the north-western two-rowed lines indicating that this locus might have an associated adaptive reward with increasing latitude.
The strongest association for early on vigor in the PPP169 console (QDEV.169.7H-ii) was found on chromosomal arm 7HS. The two most significant markers (both showing an identical pattern) at the 7H-2 QTL, 12_30894, and 12_30895, were both located within the HvFT1 locus (Supplementary Table S7). The blueprint of allelic diverseness reflected the geographical origin where both alleles were present amid the north-western lines, whereas in the south-eastern lines the allele was fixed (Supplementary Figure S2). Although, the HvCO1 cistron could non be detected in the GWAS analysis, the allele frequency for the known HvCO1 marker, BK_03 (Comadran et al., 2012), showed a similar pattern as that seen for QDEV.169.7H-ii.
Another QTL with strong association (QDEV.169.3H-1) to early vigor, heading, and maturity was detected on chromosomal arm 3HL nearby the known flowering and semi-dwarf locus, denso/sdw1, in the PPP169 panel (Figures 2, 4). The QDEV.169.3H-one had a too low allele frequency in the PPP124 panel (<0.05 MAF) to exist detected in the GWAS. The denso/sdw1 locus has been speculated to accept an consequence on both earliness and top (Kuczyńska et al., 2014), and the fixation of QDEV.169.3H-1 in the two-rowed lines might reflect the historical breeding focus to shorten culm length in Nordic two-rowed lines (Dockter and Hansson, 2015).
Two QTL, QDEV.124.2H-ane, and QDEV.124.7H-one, were associated with early vigor exclusively in the PPP124 console (Effigy 2B). QDEV.124.2H-ane included all significant markers detected on chromosome 2H in the PPP124 panel and was further confirmed by the identical allelic blueprint observed for these markers. The most pregnant marking in QDEV.124.2H-i with a position on 2HL is in shut vicinity of HvAP2, previously shown to influence tiller number and plant height (Alqudah et al., 2016; Neumann et al., 2017).
For heading (PPP169/PPP124) and maturity (PPP124) we found a QTL located nearby the locus HvCry1b (Figure iv), which plays a part in the regulation of seed dormancy (Barrero et al., 2014), and has been reported as a putative heading associated gene (Alqudah et al., 2016).
When the almost significantly associated markers for each trait were combined into allele combinations, we identified combinations with stiff outcome on earliness traits (Effigy half dozen and Supplementary Effigy S3). Especially noteworthy is the allele combination ACA which has a heat sum requirement 214 GDD below the virtually common allele combinations (GTG and GTA) in the Nordic barley cistron pool (Effigy half dozen). The ACA combination just occurred in five of the Icelandic lines (both 2-rowed and half dozen-rowed). In average, over all trials where maturity was scored, the difference in estrus sum requirement to maturity equals 13.five days shorter growth season. However, in the cool Icelandic conditions, the difference equals 19.four days less from sowing to maturity compared with the most common allele combinations. In the sub-arctic environment, where harvest is typically done in September, this time can be the difference betwixt a harvested mature crop and a crop destroyed by autumn frost and/or storms. The finding that the ACA allele combination was only constitute among Icelandic lines highlights the importance of selecting breeding lines in the target environment.
Phytohormone-Related Genes Associated With Straw Backdrop
For StL, nosotros report a QTL (QDEV.169.3H-ane) almost the denso/sdw1 locus on chromosome 3H in the PPP169 panel (Effigy 3A), with an upshot of ix cm. The GA20 oxidase gene (Hv20ox2), involved in biosynthesis of gibberellic acid (GA), has been identified as a candidate for the denso/sdw1 gene which could explain its effect on plant height (Jia et al., 2009). This QTL was, except for 6 lines, fixed amongst the two-rowed lines and therefore not detected in the GWAS analysis of the PPP124 panel (Figure 3B).
Another significant QTL (QDEV.169.2H-4), also with an effect of 9 cm, was found on 2H in the PPP169 panel. This is located close to the HvDWF11 locus (Dockter et al., 2014), which has earlier been reported every bit a brassinosteroid-related cistron in rice (Tanabe et al., 2005).
In the PPP124 panel three QTL, located on chromosomes 1H, 2H, and 5H, were institute associated with StL (Figure 3B). The meridian at 1H (QDEV.124.1H-ii) did not correlate with known loci for elevation, but Alqudah et al. (2016) found a marker but 8kb distally with an effect on tiller number. The additional QTL, QDEV.124.2H-i, and QDEV.124.5H-1, identified for StL in PPP124 were previously reported to be associated with tiller number by Neumann et al. (2017). QDEV.124.5H-1 has been reported to associate with lodging (Tondelli et al., 2013).
Interestingly, for straw breaking, several pregnant marker-trait associations within QDEV.169.2H-3 were located nearby the region of the gene HvGID2 (Figure 3; Marzec and Alqudah, 2018). In addition, a QTL, QDEV.169.5H- five, was found close to the GA20 oxidase factor HvGA20ox1 (Alqudah et al., 2016) was identified here to associate with straw breaking.
For lodging 2 QTL were establish on 5HL (Figure three), one of them, QDEV.169.5H-4, was located nearby a previously reported QTL for lodging (Tondelli et al., 2013). The relatively low significance of the associations with lodging and harbinger breaking could exist explained by the difficulty in scoring these traits as we observed very little lodging or harbinger breaking the first ii years. In year iii, the nitrogen level was doubled in Denmark and Iceland, to promote lodging, with some success in Republic of iceland just with less success in Denmark.
There was a general difference in the statistical strength of the associations between traits. Associations with earliness traits were weaker than associations with row type, early bound growth, and tiptop. Row type had by far the strongest association. Early jump growth, measured equally pinnacle of the foliage when plants had reached growth stages 31, 32, and 34 (Zadoks et al., 1974) was the 2d near significantly associated trait after row type. This trait showed a very high correlation across locations and years, had a loftier heritability, and therefore potentially a smaller number of controlling loci compared with the flowering pathway. As earliness is known to be controlled by a relatively large and intricate network of loci (Blümel et al., 2015) whereas height is controlled by few loci (Kuczyńska et al., 2013), this suggests that unproblematic inherited traits controlled by few loci were more than hands detected than circuitous traits controlled by multiple loci. These findings are therefore a validation of the GWAS model used.
Conclusion
Although, most lines in our study showed a low caste of harbinger breaking we identified i allele combination, GGA, with a significantly higher rate of straw breaking (Supplementary Effigy S3). This allele combination could be used to actively select against weak straw in the Nordic breeding programs.
The Blue distributions showed a considerable difference betwixt the 2 panels, with the PPP169 panel having a greater range of variety for all traits. This is, at to the lowest degree for the earliness traits and the early vigor, most likely due to a small number of extremely early six-rowed barley lines from Iceland, that all headed earlier than l days in the field trials (run across Supplementary Figure S1). The low number of these extremely early lines, only iii such lines were included, fabricated information technology hard to detect the effects of the underlying loci in the GWAS. Interestingly, the extremely early on lines from Iceland, all carried the same allele at markers BK_12, BK_14, BK_15, and BK_16, all located within the Ppd-H1 locus, different from the remainder of the Nordic textile. However, this allele combination was also found in the single two-rowed line of intermediate earliness from the United Kingdom. The fixed allele combination amidst the four Ppd-H1 associated markers is therefore insufficient to explain the extreme earliness observed in the Icelandic fabric, and no other loci are establish in the GWAS that could on their own explicate the extreme earliness. Evidence does, however, suggest that a polymorphism at the markers 12_30894 and 12_30895, both located within the HvFT1 locus, found in the Icelandic lines and not in the previously mentioned United kingdom line might at least partly explicate this ascertainment. To further elucidate the genetics backside the unique agronomic performances of these extremely early lines a segregating multi-parent avant-garde generation intercross (MAGIC) population has been produced.
We here report the first GWAS of developmental traits focusing exclusively on Nordic spring barley from all five Nordic countries including both two- and half dozen-rowed cultivars. Previous studies have plant Nordic barleys to carry allelic diversity in many loci affecting early heading and early maturity (Tondelli et al., 2013; Loscos et al., 2014). This was confirmed in our written report. In a few of the known flowering loci the pattern of allelic diversity is clearly different between row types, for example alleles for markers located in the HvFT1 and HvCEN genes are fixed in the two-rowed lines but there is diversity among the six-rowed lines. Based on our results we could identify platonic allele combinations for regional accommodation to the unique day length and climate conditions in the extremely northern breadth, which could help push button the margin for barley cultivation both in the north and possibly at other marginal areas.
Author Contributions
AJ, MV, MA, LR, GB, JO, BG, RH, MG, and Be were involved in the planning and experimental pattern of the study. LK, RH, RA, MI, MJ, LR, TC, MG, and JH were managing the field experiments and phenotyping in field. JO, GB, and MG were managing the laboratory experiments. TB, MG, JO, ML, GB, and JHH performed data and statistical analyses. MG, TB, JHH, and ML wrote and critically reviewed the manuscript, made the figures and finalized the tables. All authors contributed to the word of the results and the editing and approving of the final manuscript.
Funding
We give thanks NordGen and the Nordic Council of Ministers (NMR) for assistants and fiscal support of this study that was conducted within a larger project, Public Private Partnership (PPP) for Pre-breeding in barley. This piece of work was funded past NMR (No. PPP_02) and with in-kind from the private partners.
Conflict of Interest Statement
JO and AJ were employed by Nordic Seed A/S, Kingdom of denmark. TC, ST, and BG were employed by Lantmännen Lantbruk, Sweden. LR and MA were employed past Graminor, Kingdom of norway. RA, MI, and MV were employed by Boreal Constitute Convenance Ltd., Republic of finland. LK, RH, and Exist were employed past Sejet Plant Convenance, Kingdom of denmark.
The remaining authors declare that the research was conducted in the absence of whatever commercial or fiscal relationships that could exist construed as a potential conflict of interest.
Acknowledgments
Vahid Edriss (Nordic Seed A/Due south, Denmark) is gratefully acknowledged for assist with R-scripts and analyses. Áslaug Helgadóttir (Agronomical University of Iceland), Outi Manninen and Hanna Haikka (Boreal Found Breeding Ltd, Finland), Jens Due Jensen (Nordic Seed A/S, Denmark), and Stein Bergersen (Graminor, Norway) are acknowledged for their contribution to planning of the experiment and word of the results. The plant material included in the written report was provided by Nordic Seed A/Southward (Odder, Denmark), Sejet A/S (Horsens, Kingdom of denmark), Lantmännen Lantbruk (Svalöv, Sweden), Graminor A/S (Bjørke, Kingdom of norway), Boreal OI (Jokioinen, Republic of finland), and the Agricultural University of Iceland (Reykjavik, Iceland). The high throughput SNP genotyping was performed by Trait Genetics (Gatersleben, Frg). The authors wish to thank the reviewers for valuable input during the review procedure.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/x.3389/fpls.2019.00542/full#supplementary-cloth
Footnotes
- ^http://faostat3.fao.org/
- ^http://floresta.eead.csic.es/barleymap/
- ^http://taylor0.biology.ucla.edu/structureHarvester/
- ^https://github.com/YinLiLin/R-Cmplot
References
Alqudah, A. Chiliad., Koppolu, R., Wolde, G. One thousand., Graner, A., and Schnurbusch, T. (2016). The genetic architecture of barley plant stature. Front. Genet. 7:117. doi: 10.3389/fgene.2016.00117
CrossRef Full Text | Google Scholar
Baik, B.-Grand. (2016). Current and potential barley grain nutrient products. Cereal Foods World 61, 188–196. doi: 10.1094/CFW-61-5-0188
CrossRef Total Text | Google Scholar
Baik, B.-K., and Ullrich, Due south. Due east. (2008). Barley for food: characteristics, improvement, and renewed interest. J. Cereal Sci. 48, 233–242. doi: 10.1016/j.jcs.2008.02.002
CrossRef Total Text | Google Scholar
Barrero, J. Thou., Downie, A. B., Xu, Q., and Gublera, F. (2014). A function for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Constitute Cell 26, 1094–1104. doi: 10.1105/tpc.113.121830
PubMed Abstract | CrossRef Full Text | Google Scholar
Bates, D., Maechler, M., Bolker, B., and Walker, S. (2015). Fitting linear mixed effects models using lme4. J. Stat. Softw. 67, 1–48.
Google Scholar
Bengtsson, T., The PPP Barley Consortium, Manninen, O., Jahoor, A., and Orabi, J. (2017). Genetic diversity, population structure and linkage disequilibrium in Nordic spring barley (Hordeum vulgare L. subsp. vulgare). Genet. Res. Crop Evol. 64, 2021–2033. doi: 10.1007/s10722-017-0493-5
CrossRef Full Text | Google Scholar
Blümel, Chiliad., Dally, N., and Jung, C. (2015). Flowering time regulation in crops—what did we learn from Arabidopsis? Curr. Opin. Biotechnol. 32, 121–129. doi: 10.1016/j.copbio.2014.xi.023
PubMed Abstract | CrossRef Full Text | Google Scholar
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. Thou., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in various samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
PubMed Abstract | CrossRef Full Text | Google Scholar
Campoli, C., Drosse, B., Searle, I., Coupland, Thou., and von Korff, Grand. (2012). Functional characterisation of HvCO1, the Barley (Hordeum Vulgare) flowering time ortholog of CONSTANS. Plant J. 69, 868–880. doi: x.1111/j.1365-313X.2011.04839.10
PubMed Abstract | CrossRef Full Text | Google Scholar
Cantalapiedra, C. P., Boudiar, R., Casas, A. Thousand., Igartua, Eastward., and Contreras-Moreira, B. (2015). BARLEYMAP: physical and genetic mapping of nucleotide sequences and notation of surrounding loci in barley. Mol. Breed. 35:xiii. doi: 10.1007/s11032-015-0253-ane
CrossRef Total Text | Google Scholar
Chono, M., Honda, I., Zeniya, H., Yoneyama, K., Saisho, D., and Takeda, Thousand. (2003). A semidwarf phenotype of Barley Uzu results from a nucleotide commutation in the gene encoding a putative brassinosteroid receptor. Establish Physiol. 133, 1209–1219. doi: 10.1104/pp.103.026195.ii
PubMed Abstract | CrossRef Full Text | Google Scholar
Cockram, J., Jones, H., Leigh, F. J., Sullivan, D. O., Powell, West., and Laurie, D. A. (2007). Control of flowering fourth dimension in temperate cereals: genes, domestication, and sustainable productivity. J. Exp. Bot. 58, 1231–1244. doi: 10.1093/jxb/erm042
PubMed Abstruse | CrossRef Full Text | Google Scholar
Comadran, J., Kilian, B., Russell, J., Ramsay, 50., Stein, N., Ganal, M., et al. (2012). Natural variation in a homolog of antirrhinum CENTRORADIALIS contributed to bound growth habit and environmental accommodation in cultivated Barley. Nat. Genet. 44, 1388–1392. doi: 10.1038/ng.2447
PubMed Abstruse | CrossRef Full Text | Google Scholar
Dockter, C., Gruszka, D., Braumann, I., Druka, A., Druka, I., Franckowiak, J., et al. (2014). Induced variations in brassinosteroid genes define Barley acme and sturdiness, and aggrandize the green revolution genetic toolkit. Plant Physiol. 166, 1912–1927. doi: 10.1104/pp.114.250738
PubMed Abstruse | CrossRef Full Text | Google Scholar
Duggal, P., Gillanders, E. M., Holmes, T. N., and Bailey-Wilson, J. E. (2008). Establishing an adjusted p-value threshold to control the family-wide type i error in genome wide association studies. BMC genomics ix:516. doi: 10.1186/1471-2164-9-516
PubMed Abstruse | CrossRef Full Text | Google Scholar
Evanno, K., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation written report. Mol. Ecol. xiv, 2611–2620. doi: x.1111/j.1365-294X.2005.02553.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Falush, D., Stephens, M., and Pritchard, J. 1000. (2007). Inference of population construction using multilocus genotype data: ascendant markers and null alleles. Mol. Ecol. Notes 7, 574–578. doi: 10.1111/j.1471-8286.2007.01758.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Faure, S., Turner, A. South., Gruszka, D., Christodoulou, V., Davis, Southward. J., von Korff, M., et al. (2012). Mutation at the cyclic clock gene Early on MATURITY viii adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. U.S.A. 109, 8328–8333. doi: x.1073/pnas.1120496109
PubMed Abstract | CrossRef Full Text | Google Scholar
Flint-Garcia, S. A., Thornsberry, J. M., and Buckler, East. South. (2003). Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54, 357–374. doi: 10.1146/annurev.arplant.54.031902
CrossRef Full Text | Google Scholar
Ford, B., Deng, W., Clausen, J., Oliver, South., Boden, Due south., Hemming, M., et al. (2016). Barley (Hordeum vulgare) circadian clock genes can reply rapidly to temperature in an Early FLOWERING 3-dependent manner. J. Exp. Bot. 67, 5517–5528. doi: ten.1093/jxb/erw317
PubMed Abstract | CrossRef Full Text | Google Scholar
Gerland, P., Raftery, A. E., Sevcikova, H., Li, N., Gu, D., Spoorenberg, T., et al. (2014). World population stabilization unlikely this century. Science 346,234–237. doi: 10.1126/scientific discipline.1257469
PubMed Abstruse | CrossRef Full Text | Google Scholar
Gupta, P. K., Kulwal, P. L., and Jaiswal, V. (2014). "Association Mapping in Ingather Plants: Opportunities and Challenges," in Advances in Genetics, Vol. 85, eds T. Friedmann, J. Dunlap, and Due south. Goodwin (Cambridge: Academic Printing), 109–148.
Google Scholar
Hay, R. K. One thousand. (1995). Harvest index: a review of its use in plant convenance and crop physiology. Ann. Appl. Biol. 126, 197–216. doi: 10.1111/j.1744-7348.1995.tb05015.x
CrossRef Full Text | Google Scholar
Hemming, Grand. N., Peacock, Due west. J., Dennis, East. S., and Trevaskis, B. (2008). Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in Barley. Found Physiol. 147, 355–366. doi: 10.1104/pp.108.116418
PubMed Abstract | CrossRef Full Text | Google Scholar
Hilmarsson, H. Southward., Göransson, One thousand., Lillemo, Grand., Kristjánsdóttir, T. A., Hermannsson, J., and Hallsson, J. H. (2017). An overview of barley convenance and diversity trials in Republic of iceland in 1987-2014. Iceland. Agric. Sci. 30, 13–28. doi: 10.16886/IAS.2017.02
CrossRef Full Text | Google Scholar
Jia, Q., Zhang, J., Westcott, S., Zhang, X.-Q., Bellgard, G., Lance, R., et al. (2009). GA-20 Oxidase equally a candidate for the semidwarf factor sdw1/denso in Barley. Funct. Integr. Genomics ix, 255–262. doi: 10.1007/s10142-009-0120-4
PubMed Abstract | CrossRef Full Text | Google Scholar
Jones, H., Leigh, F. J., Mackay, I., Bower, M. A., Smith, L. M. J., Charles, G. P., et al. (2008). Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the fertile crescent. Mol. Biol. Evol. 25, 2211–2219. doi: 10.1093/molbev/msn167
PubMed Abstract | CrossRef Total Text | Google Scholar
Karsai, I., Szücs, P., Meszaros, Thou., Filichkina, T., Hayes, P. M., Skinner, J. S., et al. (2005). The Vrn-H2 locus is a major determinant of flowering fourth dimension in a facultative wintertime growth habit barley (Hordeum vulgare L.) mapping population. Theor. Appl. Genet. 110, 1458–1466. doi: 10.1007/s00122-005-1979-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Kastner, T., Rivas, Chiliad. J. I., Koch, W., and Nonhebel, S. (2012). Global changes in diets and the consequences for state requirements for nutrient. Proc. Natl. Acad. Sci. United statesA. 109, 6868–6872. doi: 10.1073/pnas.1117054109
PubMed Abstract | CrossRef Full Text | Google Scholar
Kuczyńska, A., Mikolajczak, K., and Cwiek, H. (2014). Pleiotrophic furnishings of the sdw1 locus in barley populations representing different rounds of recombination. Electron. J. Biotechnol. 17, 217–223. doi: x.1016/j.ejbt.2014.07.005
CrossRef Full Text | Google Scholar
Kuczyńska, A., Surma, M., Adamski, T., Mikołajczak, K., Krystkowiak, K., and Ogrodowicz, P. (2013). Effects of the semi-dwarfing sdw1/denso cistron in Barley. J. Appl. Genet. 54, 381–390. doi: 10.1007/s13353-013-0165-ten
PubMed Abstract | CrossRef Full Text | Google Scholar
Lillemo, M., Reitan, 50., and Bjørnstad, Å (2010). Increasing affect of plant breeding on barley yields in key Kingdom of norway from 1946 to 2008. Plant Breed. 129, 484–490. doi: 10.1111/j.1439-0523.2009.01710.x
CrossRef Total Text | Google Scholar
Lipka, A. E., Tian, F., Wang, Q., Peiffer, J., Li, M., Bradbury, P. J., et al. (2012). GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399. doi: 10.1093/bioinformatics/bts444
PubMed Abstract | CrossRef Full Text | Google Scholar
Lister, D. L., Thaw, S., Bower, M. A., Jones, H., Charles, K. P., Jones, One thousand., et al. (2009). Latitudinal variation in a photoperiod response gene in european barley: insight into the dynamics of agricultural spread from 'historic' specimens. J. Archaeol. Sci. 36, 1092–1098. doi: 10.1016/j.jas.2008.12.012
CrossRef Full Text | Google Scholar
Liu, H., Bayer, M., Druka, A., Russell, J. R., Hackett, C. A., Poland, J., et al. (2014). An evaluation of genotyping by sequencing (GBS) to map the breviaristatum-E (Ari-Eastward) locus in cultivated Barley. BMC Genomics fifteen:104. doi: 10.1186/1471-2164-15-104
PubMed Abstruse | CrossRef Total Text | Google Scholar
Loscos, J., Igartua, E., Contreras-Moreira, B., Pilar Gracia, G., and Casas, A. M. (2014). HvFT1 polymorphism and consequence-survey of barley germplasm and expression analysis. Front. Plant Sci. v:251. doi: 10.3389/fpls.2014.00251
PubMed Abstract | CrossRef Full Text | Google Scholar
Lundqvist, U. (2009). "80 Years of Scandinavian Barley Mutation Genetics and Breeding," in Induced Plant Mutations in the Genomics Era, ed. Q. Y. Shu (Rome: Nutrient and Agriculture System of the United Nations), 39–43.
Google Scholar
Malysheva-Otto, Fifty. V., Ganal, G. West., and Roder, M. Southward. (2006). Assay of molecular diversity, population construction and linkage genet resour ingather evol 123 disequilibrium in a worldwide survey of cultivated barley germplasm (Hordeum vulgare L. BMC Genet. 7:6. doi: 10.1186/1471-2156-7-6
PubMed Abstruse | CrossRef Total Text | Google Scholar
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544:427. doi: 10.1038/nature22043
PubMed Abstract | CrossRef Full Text | Google Scholar
Maurer, A., Draba, V., Jiang, Y., Schnaithmann, F., Sharma, R., Schumann, East., et al. (2015). Modelling the genetic architecture of flowering fourth dimension control in barley through nested association mapping. BMC Genomics 16:290. doi: x.1186/s12864-015-1459-7
PubMed Abstruse | CrossRef Full Text | Google Scholar
Neumann, Grand., Zhao, Y., Chu, J., Keilwagen, J., Reif, J. C., Kilian, B., et al. (2017). Genetic compages and temporal patterns of biomass accumulation in leap barley revealed by image analysis. BMC Constitute Biol. 17:137. doi: 10.1186/s12870-017-1085-4
PubMed Abstract | CrossRef Full Text | Google Scholar
Nitcher, R., Distelfeld, A., Tan, C. T., Yan, L., and Dubcovsky, J. (2013). Increased copy number at the HvFT1 locus is associated with accelerated flowering fourth dimension in barley. Mol. Genet. Genomics 288, 261–275. doi: ten.1007/s00438-013-0746-8
PubMed Abstruse | CrossRef Total Text | Google Scholar
Nurminiemi, M., Bjørnstad,Å, and Rognli, O. A. (1996). Yield stability and accommodation of Nordic Barleys. Euphytica 92, 191–202. doi: 10.1007/BF00022845
CrossRef Full Text | Google Scholar
Orabi, J., Jahoor, A., and Backes, G. (2014). Changes in allelic frequency over fourth dimension in European bread wheat (Triticum aestivum L.) varieties revealed using Dart and SSR markers. Euphytica 197, 447–462. doi: x.1007/s10681-014-1080-x
CrossRef Full Text | Google Scholar
Peakall, R., and Smouse, P. E. (2006). GenAlEx 6: genetic assay in excel. population genetic software for pedagogy and research. Mol. Ecol. Notes vi, 288–295. doi: 10.1111/j.1471-8286.2005.01155.10
PubMed Abstract | CrossRef Total Text | Google Scholar
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic assay in excel. population genetic software for educational activity and research–An update. Bioinformatics 28, 2537–2539. doi: x.1093/bioinformatics/bts460
PubMed Abstract | CrossRef Full Text | Google Scholar
Peng, J., Richards, D. Due east., Hartley, N. M., Potato, M. P., Devos, K. M., Flintham, J. E., et al. (1999). 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: x.1038/22307
PubMed Abstract | CrossRef Full Text | Google Scholar
R Development Core Team (2017). R: A Language and Environment For Statistical Calculating. Vienna: R Foundation for Statistical Computing.
Google Scholar
Rafalski, A. (2002). Applications of unmarried nucleotide polymorphisms in crop genetics. Curr. Opin. Institute Biol. v, 94–100. doi: 10.1016/S1369-5266(02)00240-6
CrossRef Total Text | Google Scholar
Remington, D. L., Thornsberry, J. M., Matsuoka, Y., Wilson, L. M., Whitt, S. R., Doebley, J., et al. (2001). Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. U.S.A. 98, 11479–11484. doi: 10.1073/pnas.201394398
PubMed Abstruse | CrossRef Full Text | Google Scholar
Revelle, W. (2018). Psych: Procedures for Personality and Psychological Enquiry. Evanston: Northwestern Academy.
Google Scholar
Rosenzweig, C., Elliott, J., Deryng, D., Ruane, A. C., Müller, C., Arneth, A., et al. (2014). Assessing agricultural risks of climatic change in the 21st century in a global gridded ingather model intercomparison. Proc. Natl. Acad. Sci. U.S.A. 111, 3268–3273. doi: x.1073/pnas.1222463110
PubMed Abstract | CrossRef Full Text | Google Scholar
Russell, J., Mascher, M., Dawson, I. K., Kyriakidis, S., Calixto, C., Freun, F., et al. (2016). Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet. 48, 1024–1030. doi: ten.1038/ng.3612
PubMed Abstract | CrossRef Full Text | Google Scholar
Tanabe, Southward., Ashikari, M., Fujioka, South., Takatsuto, S., Yoshida, South., Yano, M., et al. (2005). Cytochrome P450 is implicated in brassinosteroid biosynthesis via the label of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Prison cell 17, 776–790. doi: 10.1105/tpc.104.024950
PubMed Abstract | CrossRef Total Text | Google Scholar
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). From the cover: global food demand and the sustainable intensification of agronomics. Proc. Natl. Acad. Sci. U.s.a.A. 108, 20260–20264. doi: ten.1073/pnas.1116437108
PubMed Abstruse | CrossRef Full Text | Google Scholar
Tondelli, A., Xu, X., Moragues, One thousand., Sharma, R., Schnaithmann, F., Ingvardsen, C., et al. (2013). Structural and temporal variation in genetic variety of european bound two-row barley cultivars and association mapping of quantitative traits. Institute Genome 6, ane–14. doi: 10.3835/plantgenome2013.03.0007
CrossRef Full Text | Google Scholar
Turner, A., Beales, J., Faure, S., Dunford, R. P., and Laurie, D. A. (2005). The pseudo-response regulator Ppd-H1 provides accommodation to photoperiod in Barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
PubMed Abstract | CrossRef Full Text | Google Scholar
Wang, J., Yang, J., Jia, Q., Zhu, J., Shang, Y., Hua, W., et al. (2014). A new QTL for institute height in barley (Hordeum Vulgare L.) showing no negative furnishings on grain yield. PLoS One 9:e90144. doi: 10.1371/journal.pone.0090144
PubMed Abstract | CrossRef Full Text | Google Scholar
Waugh, R., Flavell, A. J., Rusell, J., Thomas, W., Ramsay, Fifty., and Comadran, J. (2014). "Exploiting Barley Genetic Resources for Genome Wide Clan Scans (GWAS)," in Genomics of Plant Genetic Resources, eds F. Sestili, Eastward. Botticella, and D. Lafiandra (Dordrecht: Springer), 238–251.
Google Scholar
Yu, J., Pressoir, G., Briggs, Westward. H., Bi, I. V., Yamasaki, M., Doebley, J. F., et al. (2006). A unified mixed-model method for clan mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. doi: 10.1038/ng1702
PubMed Abstract | CrossRef Full Text | Google Scholar
Zadoks, J. C., Chang, T. T., and Konzak, C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. doi: x.1111/j.1365-3180.1974.tb01084.x
CrossRef Full Text | Google Scholar
Zakhrabekova, South., Gough, S. P., Braumann, I., Müller, A. H., Lundqvist, J., Ahmann, G., et al. (2012). Induced mutations in cyclic clock regulator mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. U.Southward.A. 109, 4326–4331. doi: ten.1073/pnas.1113009109
PubMed Abstruse | CrossRef Full Text | Google Scholar
Zhang, Z., Ersoz, Eastward., Lai, C.-Q., Todhunter, R. J., Tiwari, H. Chiliad., Gore, Yard. A., et al. (2010). Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42, 355–360. doi: x.1038/ng.546
PubMed Abstract | CrossRef Full Text | Google Scholar
Is Genetic Makeup Or Allele Combination,
Source: https://www.frontiersin.org/articles/10.3389/fpls.2019.00542/full
Posted by: davisbantais.blogspot.com


0 Response to "Is Genetic Makeup Or Allele Combination"
Post a Comment